The cover for issue 46 of Oncotarget features Figure 6, "Establishment of a SARS-CoV-2 pseudovirus that expresses SPIKE protein variants on the envelope of a lentiviral core, infection of human airway epithelial cells or lung cancer cells, and demonstration of MEKi attenuation of infectivity on primary human cells," published in "MEK inhibitors reduce cellular expression of ACE2, pERK, pRb while stimulating NK-mediated cytotoxicity and attenuating inflammatory cytokines relevant to SARS-CoV-2 infection" by Zhou, et al. which reported that Natural Killer cells and innate-immune TRAIL suppress transformed and virally-infected cells.
In some human cells, remdesivir increases ACE2-promoter luciferase-reporter expression, ACE2 mRNA and protein, and ACE2 expression is attenuated by MEKi.
In serum-deprived and stimulated cells treated with remdesivir and MEKi we observed correlations between pRB, pERK, and ACE2 expression further supporting the role of proliferative state and MAPK pathway in ACE2 regulation.
Pseudotyped SARS-CoV-2 virus with a lentiviral core and SARS-CoV-2 D614 or G614 SPIKE protein on its envelope infected human bronchial epithelial cells, small airway epithelial cells, or lung cancer cells and MEKi suppressed infectivity of the pseudovirus.
The Oncotarget authors show a drug class-effect with MEKi to stimulate NK cells, inhibit inflammatory cytokines and block host-factors for SARS-CoV-2 infection leading also to suppression of SARS-CoV-2-S pseudovirus infection of human cells.
The Oncotarget authors show a drug class-effect with MEKi to stimulate NK cells, inhibit inflammatory cytokines and block host-factors for SARS-CoV-2 infection leading also to suppression of SARS-CoV-2-S pseudovirus infection of human cells
Dr. Wafik S. El-Deiry from The Brown University said, "Coronavirus 2 (SARS-CoV-2) infection progresses to a rapidly lethal adult respiratory distress syndrome (ARDS) associated with high mortality especially among the elderly or those with multiple comorbid conditions."
It is clear that while the host systemic inflammatory response makes patients critically ill, the host innate immune system including natural killer cells is involved in fighting and eliminating virally-infected cells.
Over the last 25 years the authors have studied this innate immune system pathway that the immune system uses to eliminate transformed and cancer cells as well as virally-infected cells.
Thus, their goal was to better understand and modulate the host immune response to increase the innate immune system early in SARS-CoV-2 infection while reducing the severe inflammation that occurs late in the disease course.
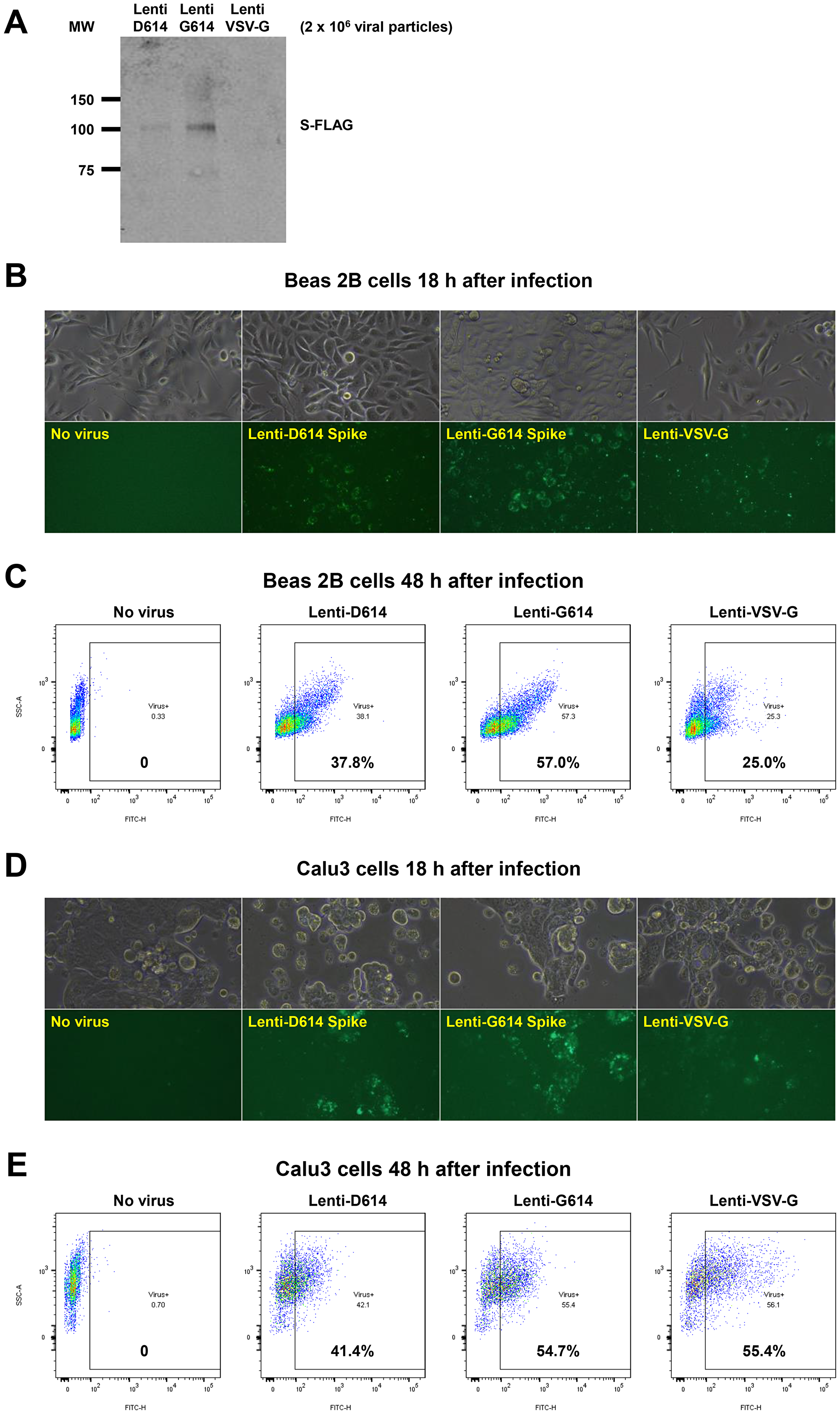
Figure 6: Establishment of a SARS-CoV-2 pseudovirus that expresses SPIKE protein variants on the envelope of a lentiviral core, infection of human airway epithelial cells or lung cancer cells, and demonstration of MEKi attenuation of infectivity on primary human cells. (A) Western blot to demonstrate FLAG-tagged S protein of SARS-CoV-2 pseudoviruses. We generated a pseudotyped SARS-CoV-2 virus which has a lentiviral core but with the SARS-CoV-2 D614 or G614 spike (S) protein on its envelope. VSV-G lentivirus was used as a negative control. Lenti- X™ p24 Rapid Titration ELISA Kit (TaKaRa) was used to determine virus titer. Equal number of lentiviral particles were analyzed on an SDS-PAGE gel followed by Western blot to detect FLAG tagged S protein. The results suggest more G614 than D614 S protein on each viral particle. (B) Fluorescence microscopic evaluation of SARS-CoV-2 pseudovirus infection of Beas 2B cells. SARS-CoV-2 pseudoviruses (5 × 106) or VSV-G lentivirus (2 × 105) were used to spin-infect Beas 2B cells in a 12-well plate. Fluorescence microscopic images were taken 18 h after infection. (C) Flow cytometry analysis of SARS-CoV-2 pseudovirus infection of Beas 2B cells. SARS-CoV-2 pseudoviruses (5 × 106) or VSV-G lentivirus (2 × 105) were used to spin-infect Beas 2B cells in a 12-well plate. Flow cytometry analysis of ZsGreen+ cells was carried out 48 h after infection. (D) Fluorescence microscopic evaluation of SARS-CoV-2 pseudovirus infection of Calu-3 cells. SARS-CoV-2 pseudoviruses (5 × 106) or VSV-G lentivirus (2 × 105) were used to spin-infect Calu-3 cells in a 12-well plate. Fluorescence microscopic images were taken 18 h after infection. (E) Flow cytometry analysis of SARS-CoV-2 pseudovirus infection of Calu-3 cells. SARS-CoV-2 pseudoviruses (5 × 106) or VSV-G lentivirus (2 × 105) were used to spin-infect Calu-3 cells in a 12-well plate. Flow cytometry analysis of ZsGreen+ cells was carried out 48 h after infection. (F) Inhibition of SARS-CoV-2 pseudovirus cell entry of human primary small airway epithelial cells. The cells were grown on 12-well plates till 80% confluence, pre-treated with the inhibitors at the indicated concentrations for 24 hr, spun-infected with the pseudoviruses followed by another 24 hr incubation with the inhibitors. All MEKi significantly blocked pseudovirus cell entry of the primary human cells while having no effect on the pantropic VSV-G lentivirus cell entry. Overall cell survival was more than 75% as determined by DAPI- in flow cytometry. The percentage of ZsGreen+ live cells was analyzed by using the FlowJo software version 10 (FlowJo, LLC, Ashland, OR).
Since remdesivir has been shown to reduce hospitalization and may reduce mortality in patients with severe COVID-19 infection, they hypothesized that suppression of viral entry into cells through inhibition of ACE2 and TMPRSS2 would reduce the spread of SARS-CoV-2 infection in a given COVID-19- patient and this would allow the innate immune system and antivirals such as remdesivir to more effectively suppress early infection.
Their results suggest that MEK inhibitors, as a class, suppress host SARS-CoV-2 infectivity factors such as ACE2 and TMPRSS2, and that alone or in combination with remdesivir, there is innate immune system activity along with suppression of inflammatory cytokines and stimulation of Natural Killer cell activity.
The El-Deiry Research Team concluded in their Oncotarget Priority Research Paper that Based on the data in this manuscript it may be reasonable to consider further preclinical experiments as well as clinical translation of the MEKi results.
Some of the open questions include a more detailed understanding of how the MAPK pathway activates ACE2, more direct evidence for effects of MEKi on actual SARS-CoV-2 infectivity of human cells, and more evidence for their effects on COVID-19 infection spread in preclinical models.
In the clinic, it may be reasonable to test MEKi such as VS-6766 or trametinib in COVID-19 infected but less severely ill patients to test the idea that MEKi could keep the infection from getting worse while allowing the body?s NK cells and innate immune mechanisms to more effectively attack virally infected cells prior to severe infection.
Consideration could be given to evaluation of MEKi ?/ antiviral agents such as remdesivir given results suggesting potentially favorable drug interactions that may allow suppression of infectivity, suppression of inflammatory cytokines, stimulation of NK cell activity, and lack of suppression of TRAIL-mediated cytotoxicity.
These effects may help antiviral agents achieve more potent disease suppression to attenuate or block COVID-19 infection that may be of use as a therapeutic approach in patients with early or less severe COVID-19 disease.
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.27799
Full text - https://www.oncotarget.com/article/27799/text/
Correspondence to - Wafik S. El-Deiry - [email protected]
Keywords - ACE2, TMPRSS2, SARS-CoV-2, COVID-19, pseudovirus
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit http://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105