Oncotarget published "Epigenetic signatures differentiate uterine and soft tissue leiomyosarcoma" which reported that leiomyosarcomas are diverse, rare, and aggressive mesenchymal soft tissue sarcomas. Epigenetic alterations influence multiple aspects of cancer, however epigenetic profiling of LMS has been limited.
These authors identified differentially methylated and differentially expressed genes associated with ULMS and STLMS using DNA methylation and RNA-seq data from primary tumors. Two main clusters were identified through unsupervised hierarchical clustering: ULMS-enriched cluster and STLMS-enriched cluster. In summary, these results indicate that differential DNA methylation and gene expression patterns are associated with ULMS and STLMS.
Dr. Nita Ahuja from The Yale University School of Medicine said, "Leiomyosarcomas (LMS) are aggressive heterogeneous mesenchymal neoplasms that account for 10–20% of soft tissue sarcomas."
LMS arise from the smooth muscle cells of different structures and organs including the uterus, retroperitoneum, abdomen, large and medium blood vessels, trunk, and extremities. Newly diagnosed patients are at high risk of distant recurrence and poor disease-specific survival. The 5-year survival rate is 42% for all stages and only 14% with distant spread, based on data from the Surveillance, Epidemiology, and End Results Program datasets. Recurrence and/or metastasis occurs in ~40% of the cases, limiting the treatment options to standard chemotherapy;however, response to first-line systemic chemotherapy is low, ranging from 5% to 33%.
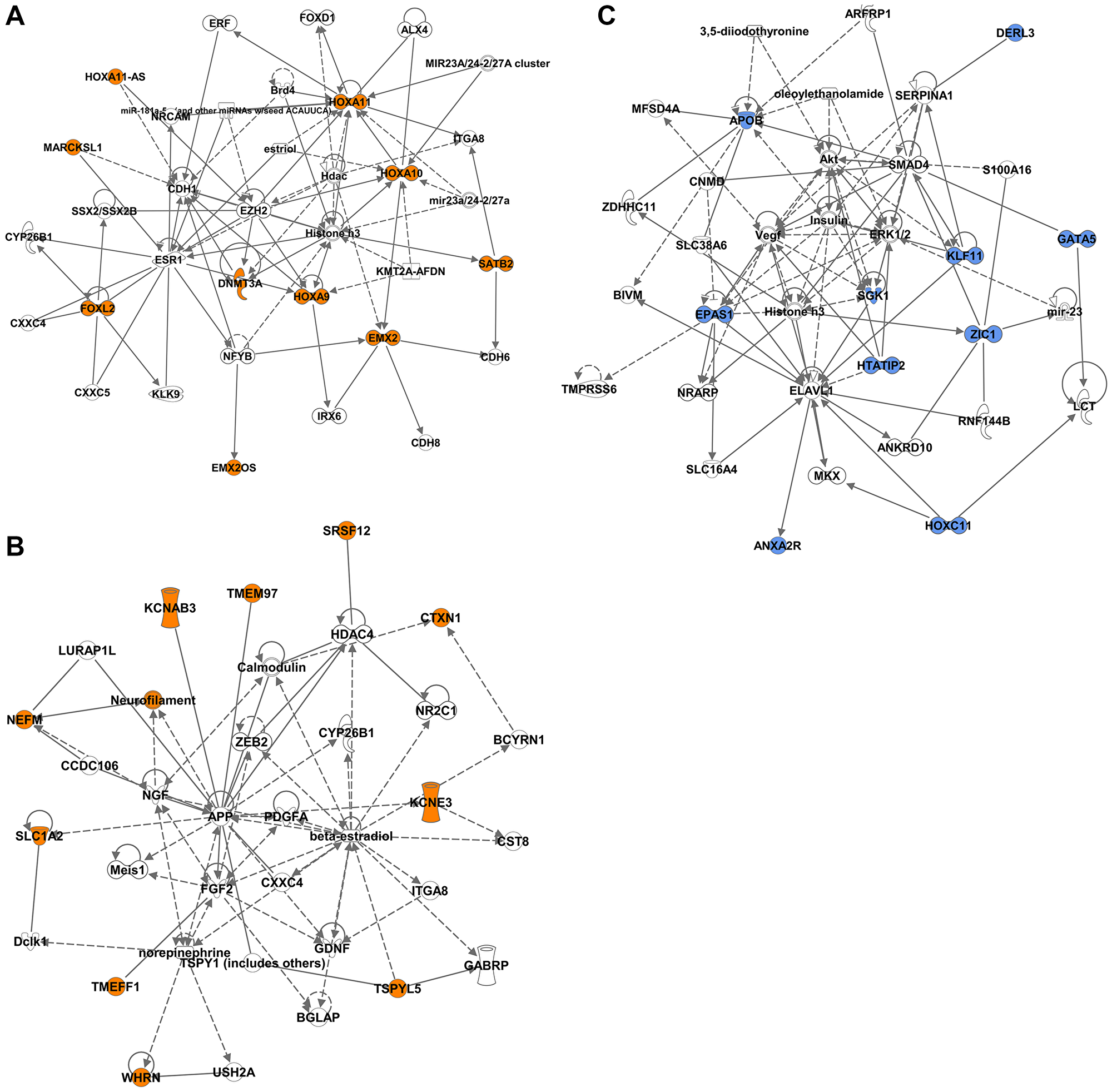
Figure 6: Network analysis of the DM-DEGs associated with STLMS-Hypermethylated-Downregulated and ULMS-Hypermethylated-Downregulated groups. Ingenuity network analysis was used to plot the gene relationships. The colored shapes indicate the downregulated genes in the STLMS-Hypermethylated-Downregulated group (orange color) (A, B) and the ULMS-Hypermethylated-Downregulated groups (blue color) (C). Genes that do not have corresponding colors, were not identified as differentially expressed in our analysis, and were integrated based on the Ingenuity Pathway Analysis evidence indicating a relevance to this network.
Current treatments for metastatic disease are either traditional chemotherapies or a targeted tyrosine kinase inhibitor. Based on the limited literature, patients with ULMS often present with larger tumors, metastatic disease, and worse overall survival. Despite this, other studies have shown that the site of origin of LMS has no impact on outcomes. Histologically ULMS and STLMS appear similar and share distinct features of the smooth muscle lineage with no available diagnostic or prognostic biomarkers to inform clinical management. Furthermore, as both subtypes are treated similarly, studies are needed to understand if molecular profiling and subtype-specific therapeutic targeting can improve survival.
Prior studies have made limited attempts to subtype LMS using exome-based, gene expression microarray-based, or RNA-seq-based profiling approaches. Epigenetic regulators are implicated in driving LMS tumor mutational heterogeneity. Furthermore, epigenetic therapies may be promising candidates for managing sarcomas and other tumor types. A previous study in their laboratory demonstrated the potential therapeutic application of hypomethylating agents such as DNA methyltransferase inhibitors in in vitro and in vivo models of LMS. These findings suggest the need for detailed epigenetic profiling of LMS tumors, exploring the roles of epigenetic alterations in oncogenesis and investigating the utility of epigenetics-based therapeutic targeting.
Epigenetic therapies may be promising candidates for managing sarcomas and other tumor types
In this study, the authors performed an integrated analysis of 98 clinically derived LMS samples and 11 controls using three publicly available datasets to identify epigenetic changes which characterize LMS subtypes and may be used as clinical biomarkers and therapeutic targets.
The Ahuja Research Team concluded in their Oncotarget Research Output, "we show evidence for differential DNA methylation profiles between ULMS and STLMS suggesting that differential epigenetic profiles are associated with LMS subtypes and may be responsible for differences seen in clinical outcomes. These findings suggest that epigenetic profiles can be used to stratify patients and apply therapeutic agents targeting the epigenetic mechanisms for LMS management. We identified several DM-DEGs and associated pathways. These findings can be used to improve our understanding of epigenetic regulation and clinical outcomes in LMS subtypes and guide biomarker development or targeted therapies."
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.28032
Full text - https://www.oncotarget.com/article/28032/text/
Correspondence to - Nita Ahuja - [email protected]
Keywords - leiomyosarcoma, epigenetics, DNA methylation, gene expression, uterine leiomyosarcoma
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit https://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105