Interview with Dr. Slack from Harvard Medical School
Oncotarget published " Cis-acting elements in its 3 ′UTR mediate post-transcriptional regulation of KRAS " which reported that the KRAS gene encodes a key signaling protein, and its messenger RNA contains an exceptionally long 3 ′UTR; this suggests that it may be subject to a highly complex set of regulatory processes.
Using extensive deletion and mutational analyses combined with luciferase reporter assays, the research team has identified inhibitory and stabilizing cis-acting regions within the KRAS 3 ′UTR that may interact with miRNAs and RNA-binding proteins, such as HuR.
Particularly, they have identified an AU-rich 49-nt fragment in the KRAS 3 ′UTR that is required for KRAS 3 ′UTR reporter repression.
In addition, they have identified another 49-nt fragment that is required to promote KRAS 3 ′UTR reporter expression.
These findings indicate that multiple cis-regulatory motifs in the 3 ′UTR of KRAS finely modulate its expression, and sequence alterations within a binding motif may disrupt the precise functions of trans-regulatory factors, potentially leading to aberrant KRAS expression.
These findings indicate that multiple cis-regulatory motifs in the 3 ′UTR of KRAS finely modulate its expression.
Dr. Frank J. Slack from The Beth Israel Deaconess Medical Center at Harvard Medical School said, " Post-transcriptional gene regulation by RNA-binding proteins (RBPs) and non-coding RNAs, such as microRNAs (miRNAs ), is critical for normal eukaryotic development and physiology. "
In these ways, 3 ′UTRs can mediate post-transcriptional gene regulation by acting as venues to coordinate interactions among various trans-regulatory factors and cis-acting 3 ′UTR elements.
Due to its exceptionally long 3 ′UTR length, the KRAS gene is presumed to be regulated at the post-transcriptional level through a highly complex interaction of cis-acting elements within its 3 ′UTR.
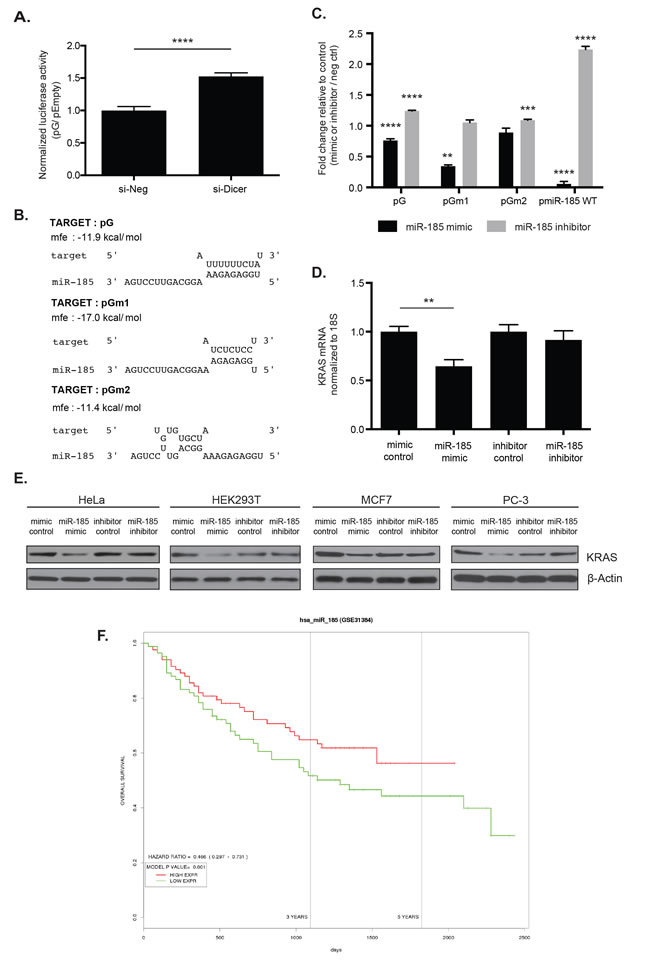
Figure 5: miR-185 potentially regulates KRAS through complementary sites within the 49-nt pG fragment. A. The pG reporter construct showed a 1.5-fold luciferase reporter de-repression in HeLa cells with global inhibition of miRNA production by Dicer knock-down (si-Dicer) compared to the control siRNA (si-Neg). ****: p -value <0.0001. B. The RNAhybrid tool predicted sequence complementarity between the seed region of miR-185 and the unmutated (G) and a T-to-C substitution mutated 49-nt fragment (Gm1). This seed region binding was abolished with a T-to-G substitution mutation (Gm2). Of note, the minimum free energy to form an RNA duplex between miR-185 and Gm1 was stronger than between miR-185 and G. C. In HeLa cells, luciferase assays showed enhanced repression with reporters pG and pGm1 with miR-185 mimic compared with the mimic control. miR-185 inhibitor induced a slight de-repression of pG and pGm2 reporter expression, compared with the inhibitor control. Overexpression and depletion of miR-185 by miR-185 mimic and inhibitor, respectively, were confirmed in HeLa cells with pmiR-185WT, which contained a perfect complementary sequence of miR-185. miR-185 mimic induced significant repression of pmiR-185WT, while de-repression was observed with miR-185 inhibitor compared with the respective controls. **: p -value <0.01; ***: p -value <0.001; ****: p -value <0.0001. D. Total RNA from HeLa cells was analyzed for KRAS mRNA level 48hrs post-miR-185 mimic or inhibitor transfection. A 35% decrease in KRAS mRNA expression was observed following miR-185 mimic transfection compared to mimic control while no change with miR-185 inhibitor. E. Total cell lysates from HeLa, HEK293T, MCF7 and PC-3 were analyzed for KRAS protein level 48hrs or 72hrs post-transfection of miR-185 mimic or inhibitor. A decrease in KRAS protein expression was observed following miR-185 mimic administration compared to mimic control in these cell lines. No change in KRAS protein levels was observed with miR-185 inhibitor. β-Actin was used as a loading control. F. The PROGmiR tool was utilized to identify a correlation between miR-185 expression and overall survival in 16 different types of cancer. High miR-185 expression was correlated with increased overall survival only in patients with liver cancer. A representative of two independent experiments is shown in mean ±S.D. in A. and D. and of three independent experiments in C.
An array of trans-regulatory factors - RBPs and miRNAs - known to regulate KRAS are often misexpressed in various types of cancer.
Through extensive deletion and mutational analyses of the KRAS 3 ′UTR, they sought to identify key cis-regulatory regions within the KRAS 3 ′UTR that interact with trans-regulatory factors.
The findings in this Oncotarget study suggest that KRAS is regulated through multiple cis-regulatory motifs in its 3 ′UTR, which have the potential to interact with various RBPs and miRNAs.
This Oncotarget study suggest that KRAS is regulated through multiple cis-regulatory motifs in its 3 ′UTR
The Slack Research Team concluded in their Oncotarget Priority Research Paper, "our findings provide evidence for the presence of multiple inhibitory and stabilizing cis-acting elements within the KRAS 3 ′UTR. Two of these elements represent individual sequence fragments with the potential to interact with post-transcriptional regulatory factors, including miRNAs and RBPs. We identified the tumor suppressive miRNA, miR-185, to interact with the KRAS 3 ′UTR via a 49-nt fragment and possibly via other regions as well, such as one miR-185 binding site about 500 bp away from the end of the 3 ′UTR predicted by TargetScan. Interestingly, within the repressive 49-nt fragment, a SNP, rs547078411, resides at the first nucleotide of the predicted miR-185 target site, and a T-to-C somatic mutation was identified at chr12: 25362140 (Hg19) in a lung cancer study (COSU583) in the COSMIC database. The potential role of these nucleotide changes in tumorigenesis remains to be determined. Further exploration to determine how multiple cis- and trans-regulatory factors collectively cooperate to regulate KRAS will provide crucial insights into the 3 ′UTR-dependent regulation of KRAS and will allow a more profound understanding of the mechanisms involved in KRAS-associated tumorigenesis. "
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.7599
Full text - https://www.oncotarget.com/article/7599/text/
Correspondence to - Frank J. Slack - [email protected]
Keywords - KRAS, 3 ′UTR, post-transcriptional regulation, microRNAs (miRNAs), miR-185
