Oncotarget published "Comparative microsomal proteomics of a model lung cancer cell line NCI-H23 reveals distinct differences between molecular profiles of 3D and 2D cultured cells" which reported that to ascertain phenotypical differences between these two distinct culture conditions, these authors carried out a comparative proteomic analysis of a membrane fraction obtained from 3D- and 2D-cultured NSCLC model cell line NCI-H23.
This analysis revealed a map of 1,166 protein species regulated in culture dependent manner, including differential regulation of a subset of cell surface-based CD molecules. They confirmed exclusive expression of CD99, CD146 and CD239 in 3D culture.
Dr. Josip Blonder from The Frederick National Laboratory for Cancer Research said, "Lung carcinoma is the deadliest cancer in the United States and worldwide."
In 2019, the American Cancer Society reported 142,670 deaths from lung cancer in the United States. Lung cancer affects men and women equally, with an overall five-year survival rate of 19%. Histologically, 80–85% of all cases belong to non-small-cell lung cancer. The most common form of NSCLC is the adenocarcinoma of the lung. Despite extensive research and economic investments, about 95% of new drugs against lung carcinoma eventually fail in clinical trials. Adherent two-dimensional in vitro cultures of human cancer cell lines are the mainstay preclinical testing model used in lung cancer drug development and discovery. In solid tumors, however, cancer cells grow in a three-dimensional environment. This makes the 2D cultured cells unable to truly reproduce the natural proliferation, migration, and/or drug permeation taking place in their innate 3D environment.
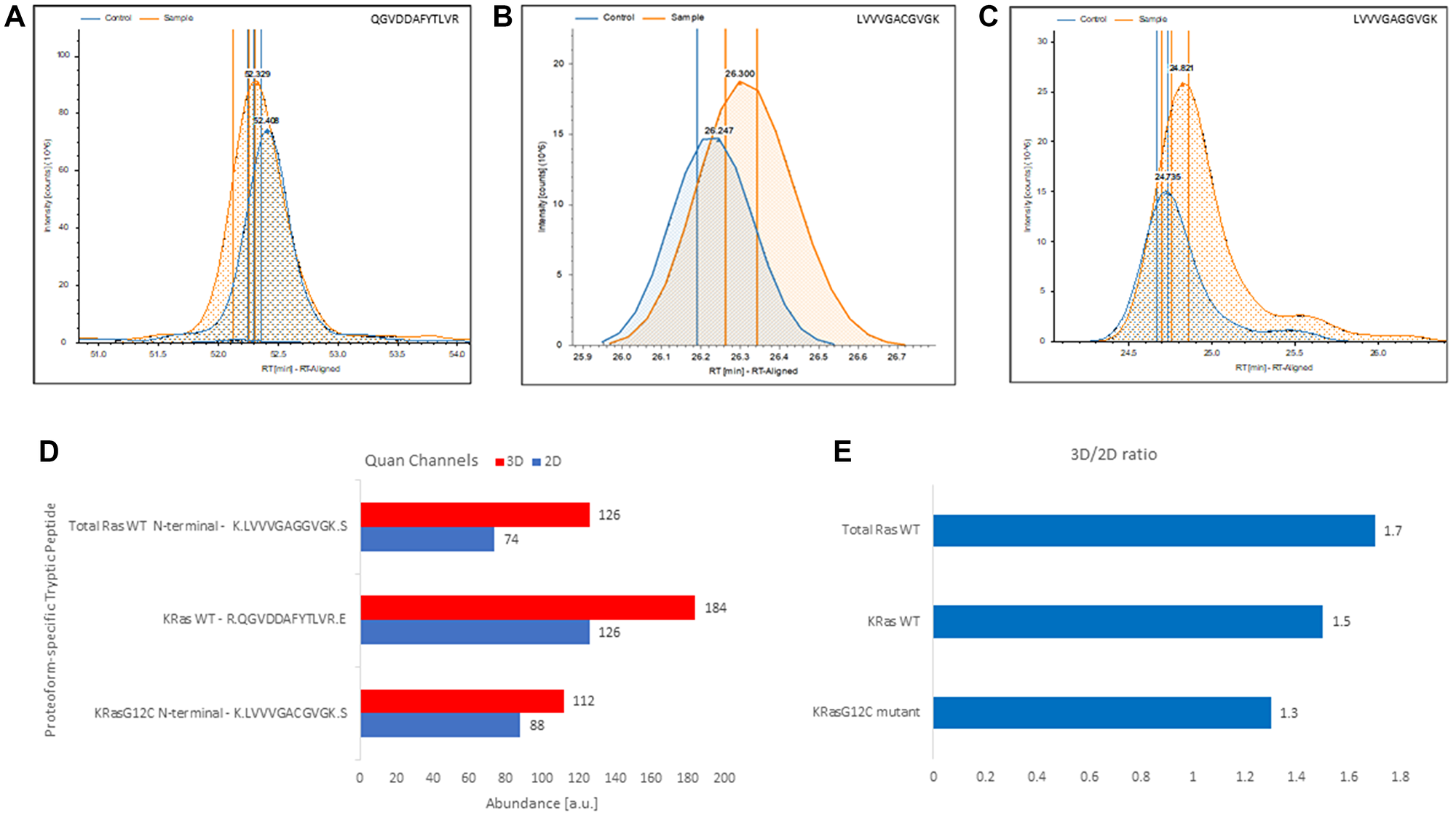
Figure 5: Label-free, gel-free, and antibody-free quantitation of Ras from complex mixture targeting exclusively proteoform-specific tryptic peptides detected in 3D-cultured and 2D-cultured NCI-H23 cells. (A) Extracted ion chromatograms of the KRas4BWT proteoform-specific tryptic peptide QGVDDAFYTLVR identified in control (i.e., 2D cultured NCI-H23 cells) and the sample (i.e., 3D-cultured NCI-H23 cells). (B) Extracted ion chromatograms of the KRas4BG12C allele-specific N-terminal mutant tryptic peptide LVVVGACGVGK identified in control (i.e., 2D cultured NCI-H23 cells) and the sample (i.e., 3D-cultured NCI-H23 cells). (C) Extracted ion chromatogram of total RasWT specific N-terminal LVVVGAVGVGK peptide identified in control (i.e., 2D cultured NCI-H23 cells) and the sample (i.e., 3D-cultured NCI-H23). (D) 3D and 2D quantitative channels readings for the total RasWT specific N-terminal LVVVGAVGVGK peptide, the KRas4BWT proteoform-specific tryptic peptide QGVDDAFYTLVR and the KRas4BG12C allele-specific N-terminal mutant tryptic peptide LVVVGACGVGK, respectively. (E) Calculated 3D/2D ratios for total RasWT, KRas4BWT, and KRas4BG12C proteoforms detected in membrane fraction of NCI-H23 cells grown in 3D and 2D culture, respectively.
To circumvent this shortcoming, the 3D cancer cell culture has been proposed as a more accurate/relevant preclinical testing model, not only biomechanically but also at the genome, proteome, and metabolome level. This approach also creates a pathway towards the development of a preclinical atlas depicting the molecular profiles of cell lines grown in 2D vs. 3D, information that is sorely needed to accurately monitor drug development. Contemporary MS-based proteomics represents a potent technology capable of revealing alterations in protein level expression/regulation and changes in post-translational modifications associated with preclinical drug evaluation.
Furthermore, the differential molecular phenotype of a NSCLC cell line membrane proteome grown in 2D- vs. 3D-culture is still missing.
The differential molecular phenotype of a NSCLC cell line membrane proteome grown in 2D- vs. 3D-culture is still missing
The Blonder Research Team concluded in their Oncotarget Research Output that the NCI-H23 cells harbors KRas4BG12C homozygous mutant and could be used as a preclinical NSCLC model for testing allele-specific covalent inhibitors that bind to the cysteine at position 12 of the G12C KRas4B mutant. Importantly, the present approach allows for direct proteoform- and allele-specific quantitation of changes in KRasWT and KRasG12C mutant regulation, and can be easily employed in quantifying responses to allele-specific covalent inhibitors or any other therapeutic compounds affecting Ras expression. The shotgun proteomics is also amenable to absolute antibody-free quantitation of Ras isoforms using synthetic heavy isoform-specific tryptic peptide standards for LC-MS-based parallel reaction monitoring, these authors previously described targeting xenotropic and polytropic retrovirus receptor 1 in a complex membrane protein mixture.
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.28072
Full text - https://www.oncotarget.com/article/28072/text/
Correspondence to - Josip Blonder - [email protected]
Keywords - lung cancer, proteomics, 2D vs. 3D cultured cells, preclinical testing model, drug target discovery
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit https://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105




