The research shows that the resulting oxadiazole-substituted aza-benzimidazole is a potent and ligand efficient S6 kinase inhibitor, which blocks the phosphorylation of RPS6 at Ser235/236 in TSC negative HCV29 human bladder cancer cells by inhibiting S6 kinase activity and thus provides a useful tool compound to investigate the function of S6 kinases.
Dr. Rob L.M. van Montfort and Dr. Keith Jones said "The 70 KDa ribosomal S6 kinases (S6K) RPS6KB1 (S6K1) and RPS6KB2 (S6K2) are key effectors of PI3K/mTOR-regulated signalling, and have been implicated in a variety of human diseases including diabetes and cancer."
S6K1, which is the most extensively studied of the two, has been shown to phosphorylate a number of substrates that regulate protein synthesis, including the 40S ribosomal protein S6, and proteins involved in translation, such as the eukaryotic initiation factor 4B and eukaryotic elongation factor 2 kinase.
In turn, S6K1 is activated by phosphorylation of the activation loop residue Thr252 by PDK1, and by phosphorylation of Thr412, located in the kinase extension region.
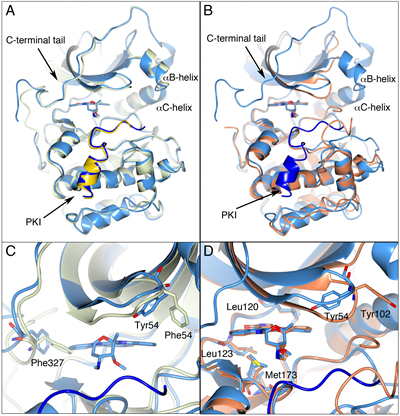
Figure 1: PKA, the PKA-S6K1 chimera and native S6K1. A) Superposition of the staurosporine-bound structures of PKA (PDB code: 1STC) in light green and the PKA-S6K1 chimera in light blue showing the close similarity of the structures. The inhibitor peptide PKI is shown in yellow and blue in the respective structures. The staurosporine molecules are shown in cylinder representation. Relevant secondary structure elements are labelled. B) Superposition of the staurosporine-bound structures of the PKA-S6K1 chimera in light blue and native S6K1 (PDB code: 3A62) in orange showing the similarity in their tertiary structures and the nearly identical binding mode of staurosporine. The inhibitor peptide PKI present in the PKA-S6K1 chimera structure is shown in blue. C) Close-up of the superposition of the staurosporine bound PKA-S6K1 chimera and the adenosine-bound PKA structure (PDB code: 1BKX) which represents an intermediate conformation in the PKA catalytic cycle. The colour scheme is the same as in panel A, but the adenosine molecule bound in 1BKX is not displayed for clarity. Phe327 in the C-terminal tail and the aromatic residue at the tip of the P-loop (Tyr54/Phe54) adopt different conformations in the respective structures. D) Close-up of the S6K1 and PKA-S6K1 chimera superposition. The positions of four of the five mutations in the PKA-S6K1 chimera ATP-binding site (Tyr54, Leu120, Leu123, Met173) are highlighted using the PKA-S6K1 sequence numbering. The fifth mutation near the ATP-binding site (Lys181) could not be shown in this orientation. Also shown is Tyr102 in S6K1. All structural figures were made using CCP4MG [48].
However, for full activation of S6K1, these phosphorylation events have to be preceded by phosphorylation of a series of serine and threonine residues in the C-terminal autoinhibitory domain and by phosphorylation of Ser394 in the turn-motif.
Studies in cultured cells have confirmed that S6K activity is enhanced by mechanisms activating the PI3-kinase/Akt/mTOR pathway, such as loss of the tumour suppressor PTEN, and have shown a positive correlation between S6K activity and tumour growth.
Finally, increased phosphorylation of S6K and corresponding phosphorylation of RPS6 was observed in HPV16-infected cervical cancer tissue samples, and the constitutive activation of S6K1 is associated with cisplatin resistance in human H69 small cell lung cancer cells.
The Montfort/Jones Research Team concluded, "in addition, we developed a novel robust crystallography system based on a PKA-S6K1 chimera and experimentally determined the binding mode of two of the three inhibitor series, including the improved and cellularly active azabenzimidazole inhibitor."
Full text - https://doi.org/10.18632/oncotarget.1255
Correspondence to - Rob L.M. van Montfort - [email protected] and Keith Jones - [email protected]
Keywords - S6 kinase, P70S6K, cancer, inhibitor, structure-based drug design