The cover for issue 52 of Oncotarget features Figure 1, "Volcano plots of DNA methylation in tumor tissues compared with nontumor tissue," published in "Reduction of T-Box 15 gene expression in tumor tissue is a prognostic biomarker for patients with hepatocellular carcinoma" by Morine, et al. which reported that the authors conducted a genome-wide analysis of DNA methylation of the tumor and non-tumor tissue of 15 patients with Hepatocellular carcinoma (HCC), and revealed TBX15 was the most hypermethylated gene of the tumor.
Another validation set, which comprised 58 HCC with radical resection, was analyzed to investigate the relationships between tumor phenotype and TBX15 mRNA expression.
TBX15 mRNA levels in tumor tissues were significantly lower compared with those of nontumor tissues.
Multivariate analysis identified low TBX15 expression as an independent prognostic factor for overall and disease-free survival.
Therefore, genome-wide DNA methylation profiling indicates that hypermethylation and reduced expression of TBX15 in tumor tissue represents a potential biomarker for predicting poor survival of patients with HCC.
Reduced expression of TBX15 in tumor tissue represents a potential biomarker for predicting poor survival of patients with HCC
Dr. Yuji Morine from the Tokushima University Graduate School said, "Hepatocellular carcinoma (HCC) is the sixth most prevalent human cancer and third-highest cause of cancer-related deaths worldwide, and its incidence is significantly increasing."
Several genome-wide studies of the molecular alterations in tumor and non-tumor liver tissues addressed DNA, mRNA, and microRNA expression as well as epigenetic alterations to predict recurrence and hepatocarcinogenesis after treating patients with curative intent according to distinct gene expression or alteration patterns.
Here the Oncotarget authors focused on the epigenetic alterations of the genomes of the tumor and non-tumor cells of patients with HCC to identify the mechanisms of tumor progression and hepatocarcinogenesis because of global DNA hypomethylation or cancer specific DNA hypermethylation occurs in certain carcinomas.
They firstly chose long interspersed nuclear element-1 sequences that provided a surrogate marker of global DNA methylation levels and detected hypomethylation of LINE-1 in HCC tissues, which was significantly associated with poor prognosis of patients with HCC through the activation of MET.
Further, they investigated epigenetic characteristics with array-based analysis of DNA methylation using the Illumina Human Methylation 450 BeadChip and identified specific DNA methylation profiling in nontumor liver tissue of patients without HCV and HBV detectable infection, which possibly contributed to the development of HCC.
In this study, they analyzed DNA methylation profiles in tumor tissue of HCC using those case series of their recent study, and found the most hypermethylated gene in tumor compared with nontumor tissues, encodes T-box 15, implicating TBX15 as a candidate regulator of tumor progression.
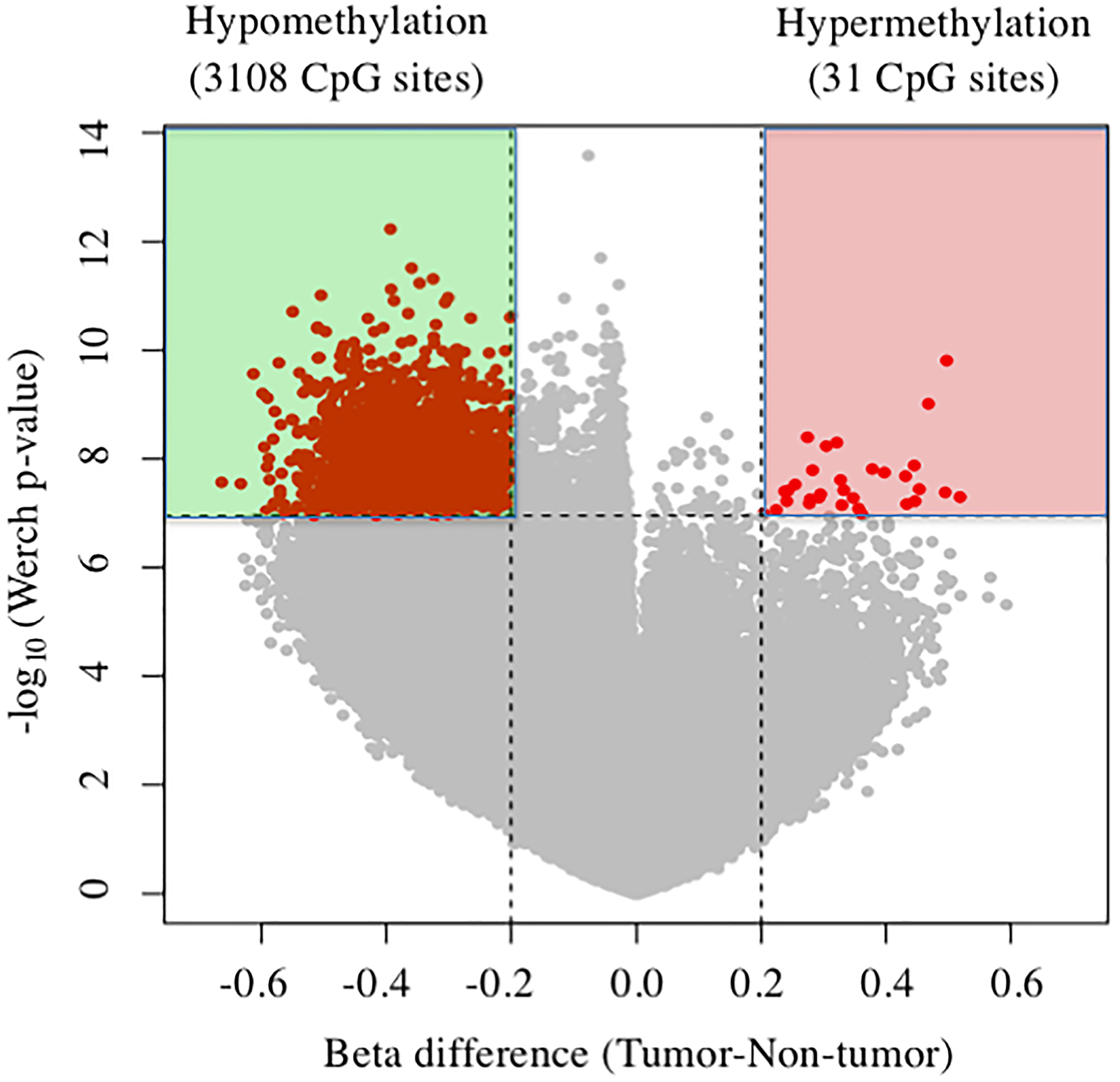
Figure 1: Volcano plots of DNA methylation in tumor tissues compared with nontumor tissue. Significant associations are indicated in red.
The Morine Research Team concluded in their Oncotarget Research Paper that together, the present and previous findings support the importance of conducting further investigations to determine the role of TBX15 in cancer, particularly in tumor progression.
Second, selection bias was possible, because of the retrospective one.
Third, the authors did not determine the actual mechanism of TBX15 for tumor malignancy in a basic experiment.
Those issues should be solved in several cancers, to clarify the true function of TBX15 for tumor malignancy in each cancer.
In conclusion, reduced expression of TBX15 may serve as a potential biomarker for predicting tumor progression and poor survival as well as a target for antitumor therapy in HCC.
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.27852
Full text - https://www.oncotarget.com/article/27852/text/
Correspondence to - Yuji Morine - [email protected]
Keywords - genome-wide analysis, methylation, tumor suppressor gene, prognostic biomarker, hepatocellular carcinoma
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit http://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105