Oncotarget published "Identification of nuclear export signal in KLLN suggests potential role in proteasomal degradation in cancer cells" which reported that on KLLN localization in both the nucleus and cytoplasm and the identification of a putative nuclear export signal sequence.
Inhibition of the CRM1 export pathway increased nuclear sequestration of KLLN, confirming the prediction of an NES sequence.
Point mutations introduced in the predicted NES sequence decreased the strength of the NES and increased the nuclear sequestration of KLLN.
Contrary to expectations, the transcription regulation and cellular proliferation functions of KLLN were unaffected by increased KLLN nuclear sequestration.
Instead, increased nuclear KLLN correlated with increased nuclear sequestration of TRIM25 and decreased inhibitory phosphorylation of MDM2. Computational analysis of The Cancer Genome Atlas dataset showed positive correlation among KLLN, TRIM25 and MDM2 expression; pathway analysis of the common genes downstream of these three genes revealed protein degradation as one of the top canonical pathways.
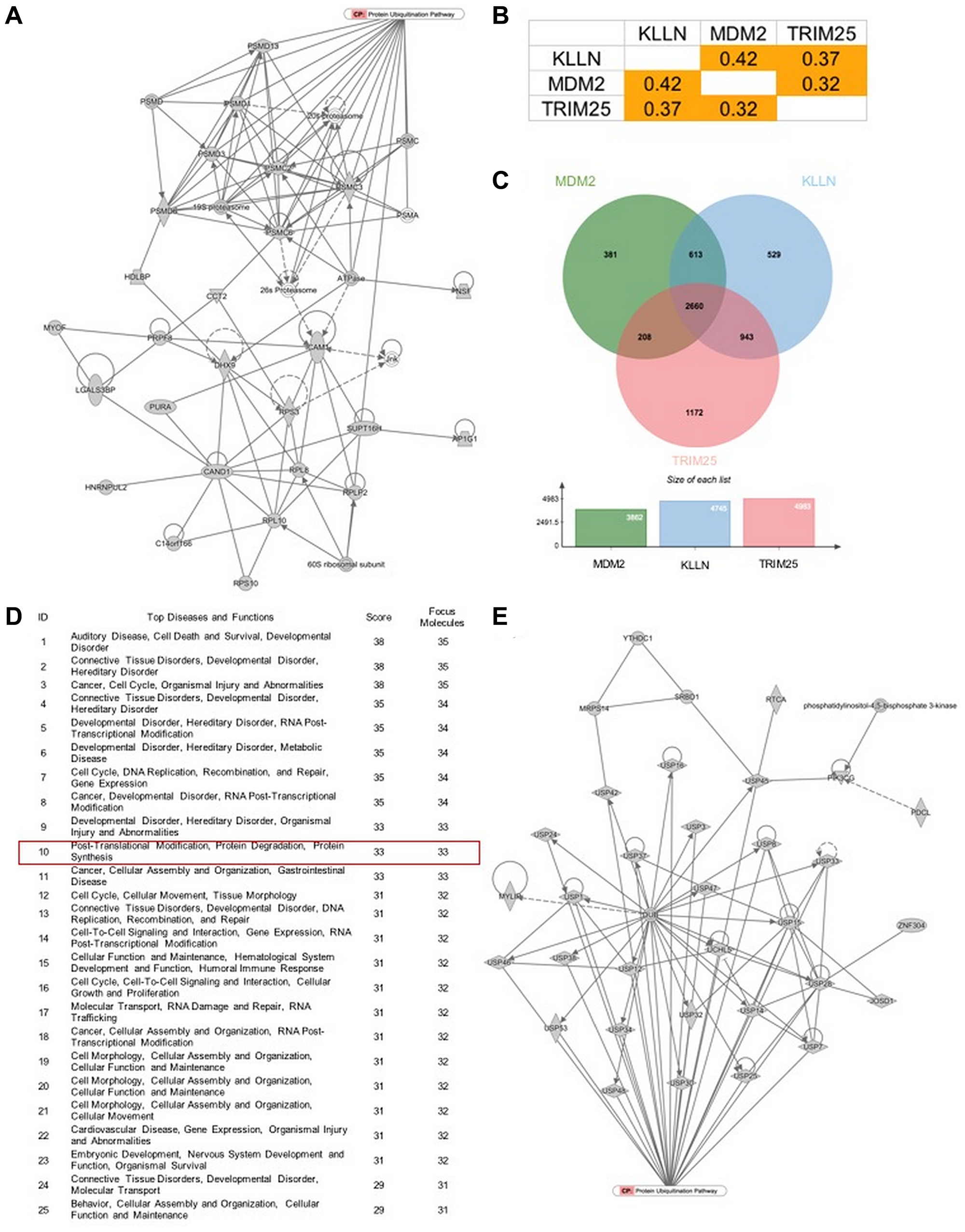
Figure 5: KLLN function correlates with proteasomal degradation. (A) IPA-based pathway analysis image of direct interactors of KLLN identified by mass spectrometry involved in protein degradation pathway. (B) Positive correlation of gene expression of KLLN, TRIM25 and MDM2 in renal cell carcinoma data collected from TCGA. (C) A Venn diagram of positively correlated genes associated with KLLN, TRIM25 and MDM2 show more than half of positively-correlated genes were common to KLLN, TRIM25 and MDM2. (D) IPA-based pathway analysis showing protein degradation as a top 10 pathway associated with the common genes positively correlated with KLLN, TRIM25 and MDM2. (E) Graphical representation of the contribution of the common positively correlated genes in protein degradation/proteasomal degradation.
Dr. Charis Eng from The Cleveland Clinic as well as Case Western Reserve University said, "KLLN is a tumor suppressor protein discovered in 2008 when researchers were searching for potential new targets of p53 that were involved in S-phase cell cycle checkpoint regulation."
In PTEN mutation negative CS/CSL, KLLN germline promoter hypermethylation is observed in up to 35% of patients and is associated with three-fold increased prevalence of breast carcinomas and two-fold increased prevalence of renal cell carcinomas.
Evidence supporting somatic KLLN deletions have also been observed in 21% of breast carcinomas in The Cancer Genome Atlas and there has been an established association between increased tumor grade and decreased KLLN expression in breast carcinomas versus adjacent normal tissue.
KLLN is transcriptionally regulated by p53 and has known p53-binding sites on its promoter, that when masked by the promoter hypermethylation, results in decreased KLLN expression.
Both H3K9 trimethylation and H3K9 methyltransferase activity have been shown to be positively correlated with KLLN expression, while maintenance of pericentric heterochromatin and genetic stability negatively correlated with KLLN expression.
In this Oncotarget study, the authors show that KLLN possesses a nuclear export signal that helps it exit the nucleus and that the likely role KLLN plays in trafficking proteins between the nuclear and cytoplasmic compartments may relate to proteasomal degradation.
In this Oncotarget study, the authors show that KLLN possesses a nuclear export signal that helps it exit the nucleus and that the likely role KLLN plays in trafficking proteins between the nuclear and cytoplasmic compartments may relate to proteasomal degradation.
The Eng Team concluded in their Oncotarget Research Output, "our results identifying the putative NES sequence of KLLN establishes KLLN as a cargo protein for the CRM1 export pathway complex. Further, the association between KLLN localization and proteasomal degradation establishes a potential new function for KLLN. Even though TRIM25 is a known regulator of p53 and MDM2 expression and activity, our study establishes a correlative role for KLLN in this pathway. In our previous study, we had suggested the closing of a regulatory loop between p53 and KLLN in response to DNA damage and this study has contributed the missing pieces to that regulatory loop."
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.27837
Full text - https://www.oncotarget.com/article/27837/text/
Correspondence to - Charis Eng - [email protected]
Keywords - KLLN, nuclear export, NES, proteasomal degradation
About Oncotarget
Oncotarget is a bi-weekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit https://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105