The cover for issue 44 of Oncotarget features Figure 1, "Schematic diagram of the human IGF2 gene structure," by Radhakrishnan, et al. recently published in "Methylation of a newly identified region of the INS-IGF2 gene determines IGF2 expression in breast cancer tumors and in breast cancer cells" which reported that these authors previously demonstrated that IGF2 protein levels are higher in BC tissues from African American women than in Caucasian women.
They also showed that high IGF2 protein levels are expressed in normal breast tissues of African American women while little or no IGF2 was detected in tissues from Caucasian women.
Thus, they designed this study to determine if differentially methylated regions of the IGF2 gene correspond to IGF2 protein expression in paired breast tissues and in BC cell lines.
The Oncotarget authors propose that methylation of DVDMR represents a novel epigenetic biomarker that determines the levels of IGF2 protein expression in breast cancer.
"The Oncotarget authors propose that methylation of DVDMR represents a novel epigenetic biomarker that determines the levels of IGF2 protein expression in breast cancer."
Since IGF2 promotes metastasis and chemoresistance, we propose that IGF2 levels contribute to BC aggressiveness.
Dr. Daisy D. De León from The Loma Linda University School of Medicine said, "Insulin-Like Growth factor 2 (IGF2) is a fetal growth factor that plays a critical function in fetal differentiation and metabolism by signaling through the IGF-I receptor and the insulin receptor."
Thus, IGF2 expression is important in normal breast development and increased IGF2 expression in the mammary gland contributes to BC malignancy.
This gene is located on the short arm of chromosome 11 at position 15.5. Methylation of the IGF2 gene regulatory regions occurs during the formation of an egg or sperm cell, and it is distinct and differentially modified depending on the parental origin of the allele.
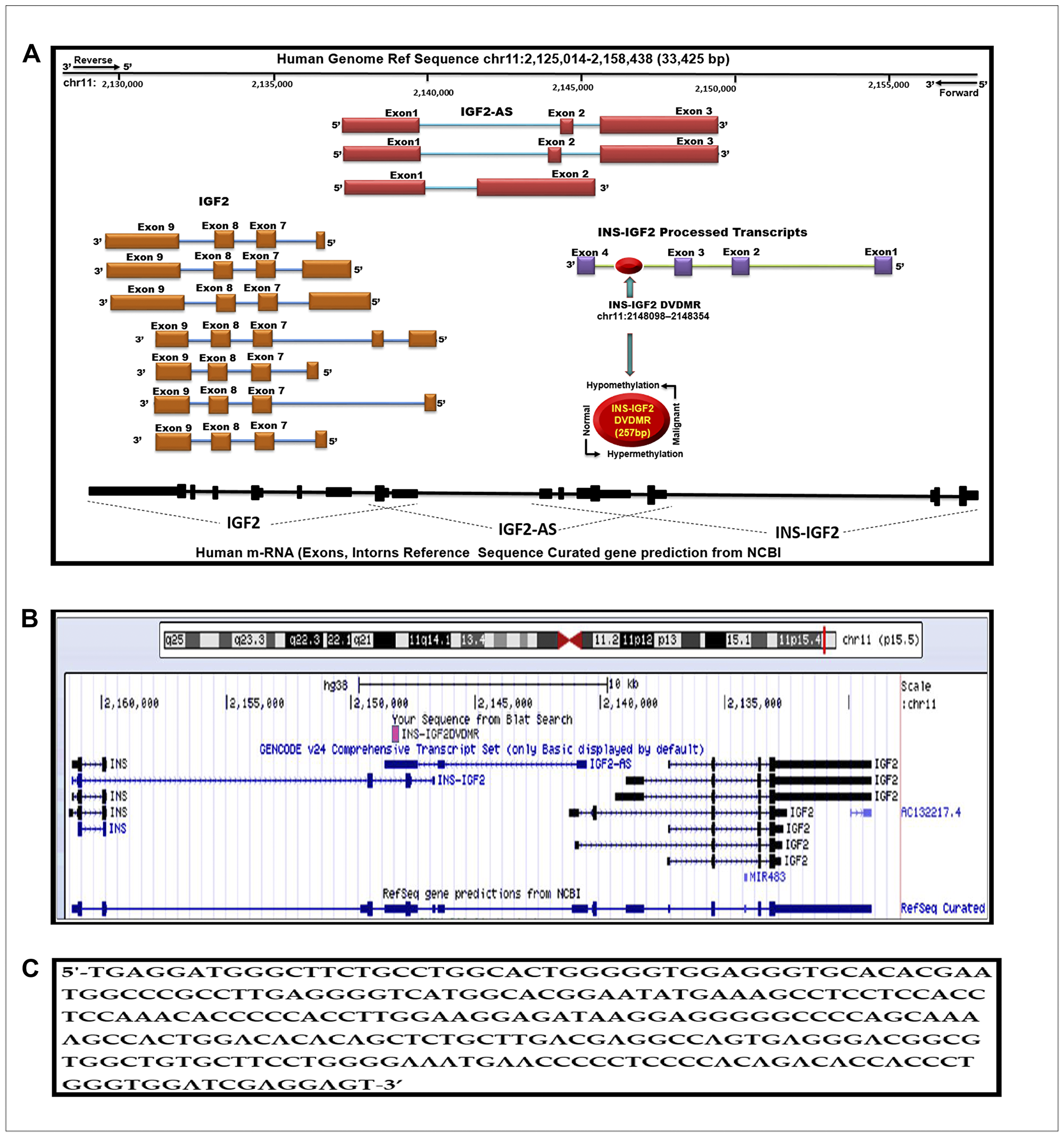
Figure 1: Schematic diagram of the human IGF2 gene structure. (A) The IGF2 gene diagram showing the relative positions of the IGF2 introns and coding exons. Also present in this region is the INS-IGF2 and the IGF2AS genes which code for non-translatable mRNAS. The newly identified differentially methylated region (DVDMR) is represented in red (chr11:2148098-2148354). This region is hypermethylated in normal breast tissues and hypomethylated in breast cancer. (B) Figure obtained from the UCSC Genome Browser, Dec. 2013 (GRCh38/hg38) showing in pink the DVDMR located between exon 1 and exon 2 of the IGF2 gene showing chr11:2148098 2148354 5′-3′ pad strand = + sequence from Blat search in reference to human genome sequence; (C) INS-IGF2 DVDMR BLAT search sequence (chr11:2148098-2148354).
In particular, dysregulation in the methylation of the IGF2 gene promoters occurs in several cancers including BC and this altered methylation leads to different clinical features in BC disease.
In spite of these advances, there is currently no consensus regarding the methylation status of the IGF2 gene, and its relationship to the levels of IGF2 protein expressed in normal breast or in breast cancer tissues.
DNA methylation patterns of the IGF2 gene were also analyzed in several BC cell lines to determine if there was a correlation between methylation of the IGF2 gene regulatory regions and the cellular expression levels of IGF2 protein.
The De León Research Team concluded in their Oncotarget Research Paper, "the present study shows that IGF2 expression in BC cells and in paired Normal- Malignant breast tissues are determined by the methylation of a novel region in the INS-IGF2 locus. We are currently studying the mechanisms underlying the methylation of the INS-IGF2 and how they control IGF2 expression. Upregulation of IGF2 in terms of the methylation patterns of the DVDMR may have an important function in the tumorigenesis of the breast. In conclusion, we propose that the INS-IGF2 DVDMR may be a useful tool to identify women at risk of developing a more aggressive BC disease."
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.27655
Full text - https://www.oncotarget.com/article/27655/text/
Correspondence to - Daisy D. De León - [email protected]
Keywords - IGF2, INS-IGF2, hypermethylation, DMR, epigenetics
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit http://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105