Oncotarget published "Dishevelled-1 DIX and PDZ domain lysine residues regulate oncogenic Wnt signaling" which reported that K69 and K285 regulate binding of DVL-1 to Wnt target gene promoters and modulate expression of Wnt target genes including CMYC, OCT4, NANOG, and CCND1, in cell line models and xenograft tumors.
Finally, these authors report that conserved DVL-1 lysines modulate various oncogenic functions such as cell migration, proliferation, cell-cycle progression, 3D-spheroid formation and in-vivo tumor growth in breast cancer models. Collectively, these findings highlight the importance of DVL-1 domain-specific lysines which were recently shown to be acetylated and characterize their influence on Wnt signaling.
these findings highlight the importance of DVL-1 domain-specific lysines which were recently shown to be acetylated and characterize their influence on Wnt signaling
Dr. Kevin Pruitt from The Texas Tech University Health Sciences Center said, "Wnt signaling is integral for normal development and tissue homeostasis."
With 19 Wnt ligands, 10 Frizzled receptors and 3 Dishevelled proteins participating in signaling, the amount of information relayed is enormous. The canonical or β-catenin dependent branch of Wnt signaling relays information through secreted Wnt ligands, transmembrane Frizzled receptors, low-density lipoprotein receptor related protein 5/6 , DVL proteins, and active β-catenin to initiate transcription of Wnt target genes.
DVL integrates and transmits complex Wnt signals, yet how it conducts this symphony of activity remains unclear. Regulation by various PTMs, however, have been shown to play a critical role specifying how the molecular signals are routed. Some of the well-studied PTMs of DVL include phosphorylation, ubiquitination, and methylation. These results indicate that unlike K69 and K285, K34 does not control nuclear localization of DVL-1 across multiple cell line models. For the first time they demonstrate that all 3 DVL-1 residues differentially regulate canonical Wnt signaling. However, the K34 site is particularly potent in regulating active β-catenin levels and total β-catenin sub-cellular localization.
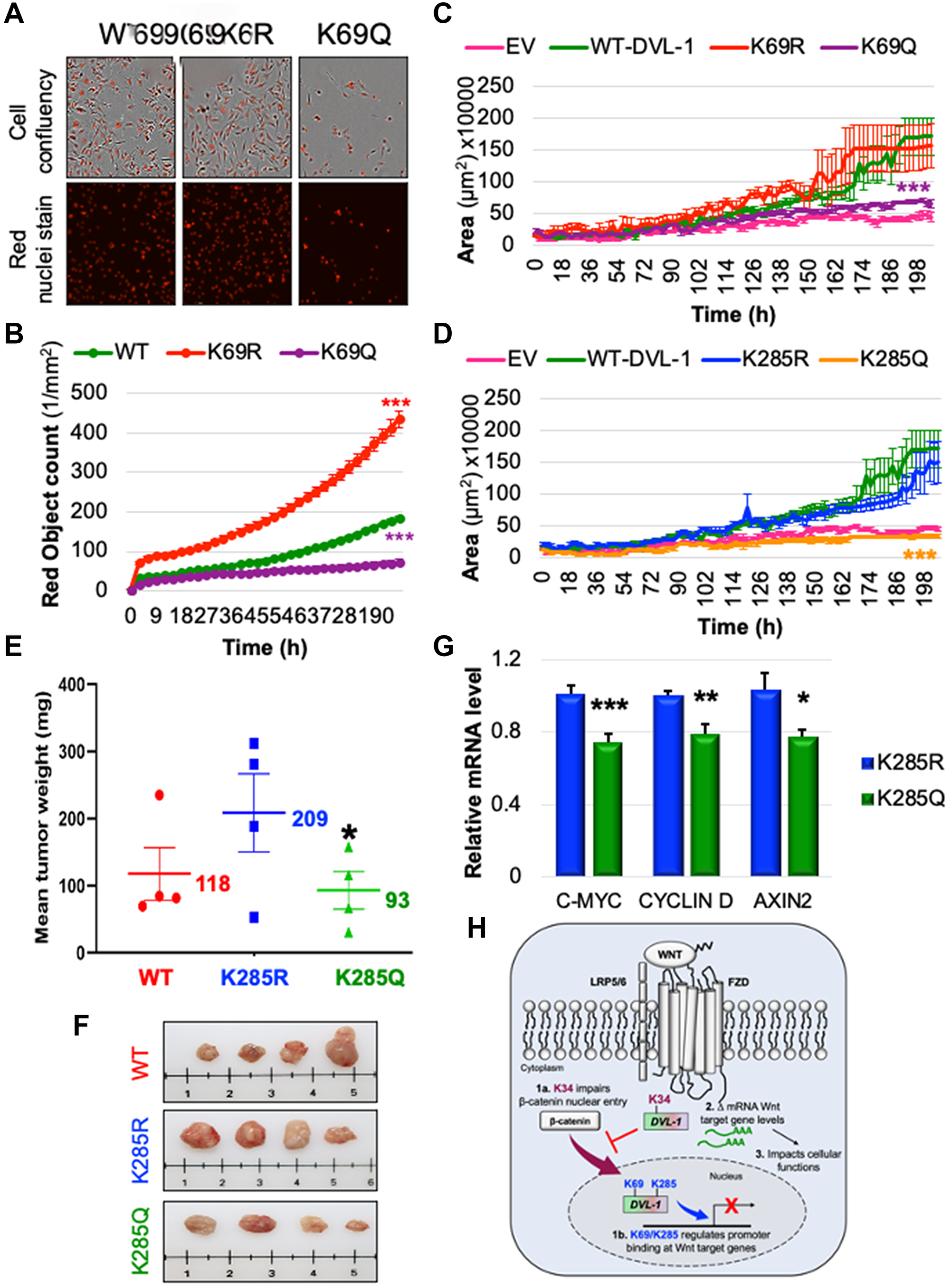
Figure 6: K69 and K285 residues significantly influence rate of cell proliferation and xenograft tumor growth respectively in MDA-MB-231 cells. (A) MDA MB-231 cells were seeded onto 96 well plates and grown in the presence of nuclear stain NucLight Rapid Red Reagent (Essen Biosciences). Cells were monitored and stained nuclei were quantified in real time by live cell imaging, with images captured every 2 hours for 120 hours. Images shown here represent confluency (upper panel) versus nuclei stain (bottom panel). (B) Each dot represents the nuclear counts at each time point. Data are shown as the mean nuclear counts (red fluorescent bodies/mm2) ± SEM and considered significant at ***p < 0.001 compared to WT-DVL1. Spheroid formation assay was conducted in MDA-MB-231 cell lines stably expressing DVL-1 (C) K69 and (D) K285 mutants. Quantification of the area of spheroids is presented as the mean ± SD, where n ≥ 5 showing statistical difference after 200h ***p < 0.001. (E) Mean tumor weight of WT-DVL1, K285R and K285Q MDA-MB-231 cells subcutaneously injected in mammary fat pad of immunocompromised mice (n = 4 per group, “*” represents significant change between R and Q mutants where *p < 0.05). (F) Representative images for tumors from mice receiving WT-DVL1, K285R and K285Q MDA-MB-231 cells (top to bottom). (G) mRNA expression of WNT target genes such as CMYC, CYCLIND1, and AXIN2 was determined by qRT-PCR (mean ± SEM, normalized to β-actin as control) using tumor obtained from xenograft studies, where n = 4. The sign “*” represents significant change in gene expression. All results are expressed as mean ± SEM and considered significant at *p < 0.05, **p < 0.01 and ***p < 0.001. (H) Schematic representation of the functional significance of lysine residues on the DIX and PDZ domains od DVL-1 with respect of Wnt signaling. While highly conserved K34 residue is critical for the entry of β-catenin into nucleus, K69/K285 residue regulate DVL-1 binding to promoter binding at Wnt target genes, and repressing their expression thereby impact broad cellular functions.
The Pruitt Research Team concluded in their Oncotarget Research Output that early discoveries associated the phenotype of DVL with disorientation in Drosophila wing hair and segment polarity in Xenopus embryo, and social abnormalities in mice. Interestingly, their functional assays conducted in cancer models demonstrate that key lysine residues subject to acetylation play a critical role in modulating cell migration, proliferation, cell cycle progression, and in vivo tumor growth, further bolstering the concept that these novel DVL-1 PTMs are functionally significant and could impact Wnt signaling branches and multiple cellular processes linked with tumorigenesis.
Sign up for free Altmetric alerts about this article
DOI - https://doi.org/10.18632/oncotarget.28089
Full text - https://www.oncotarget.com/article/28089/text/
Correspondence to - Kevin Pruitt - [email protected]
Keywords - dishevelled (DVL), post-translational modification, lysine residue, gene expression, chromatin immunoprecipitation (ChIP)
About Oncotarget
Oncotarget is a biweekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https://www.oncotarget.com or connect with:
SoundCloud - https://soundcloud.com/oncotarget
Facebook - https://www.facebook.com/Oncotarget/
Twitter - https://twitter.com/oncotarget
LinkedIn - https://www.linkedin.com/company/oncotarget
Pinterest - https://www.pinterest.com/oncotarget/
Reddit - https://www.reddit.com/user/Oncotarget/
Oncotarget is published by Impact Journals, LLC please visit https://www.ImpactJournals.com or connect with @ImpactJrnls
Media Contact
[email protected]
18009220957x105